Code, input and output for reproducing RNAseq images in manuscript
20240208_095535_DGE.rds
0 → 100644
File added
Day4_volcano_DEGs.png
0 → 100644
370 KB
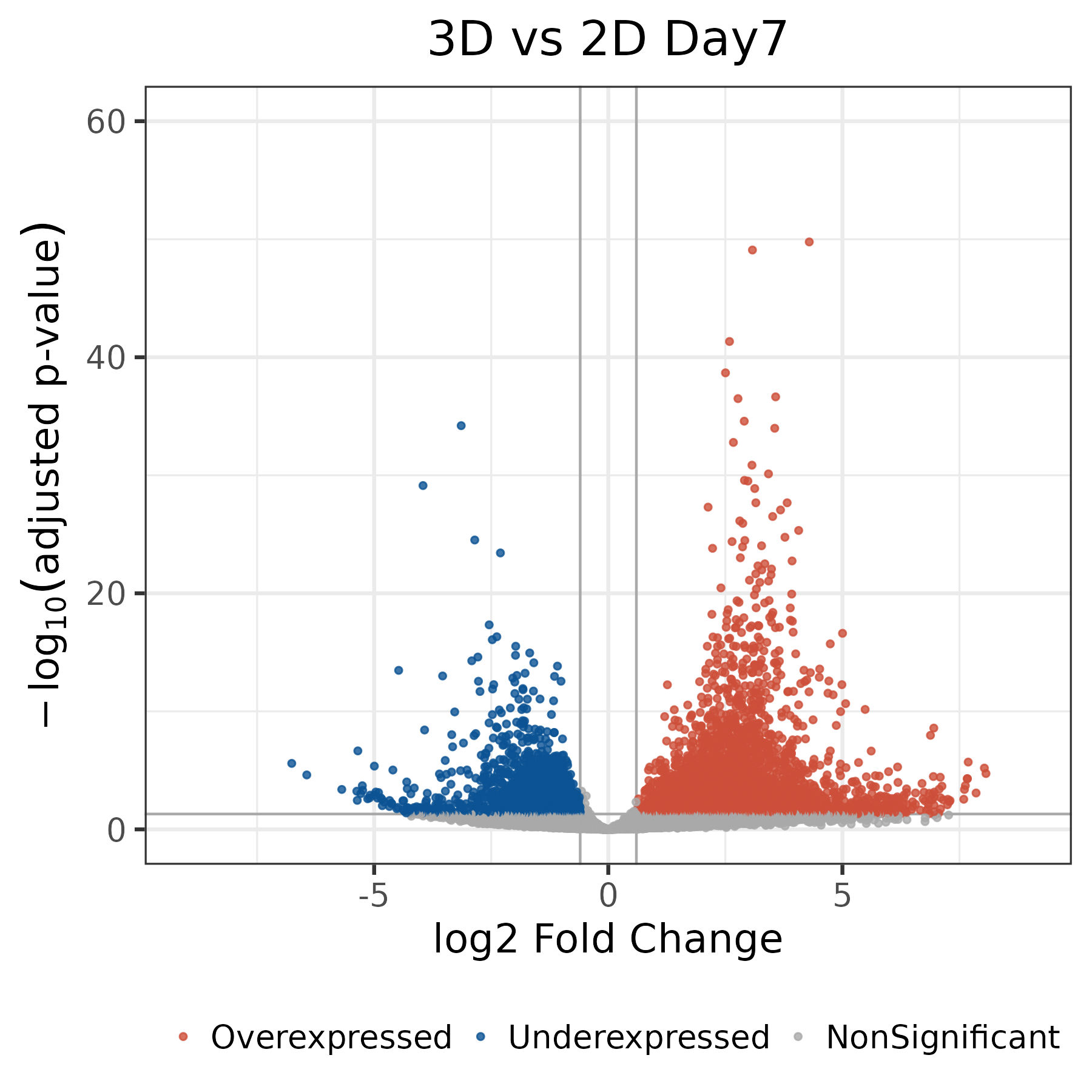
Day7_volcano_DEGs.png
0 → 100644
327 KB
DotPlotVolcano.R
0 → 100644
GOI_dotplot.png
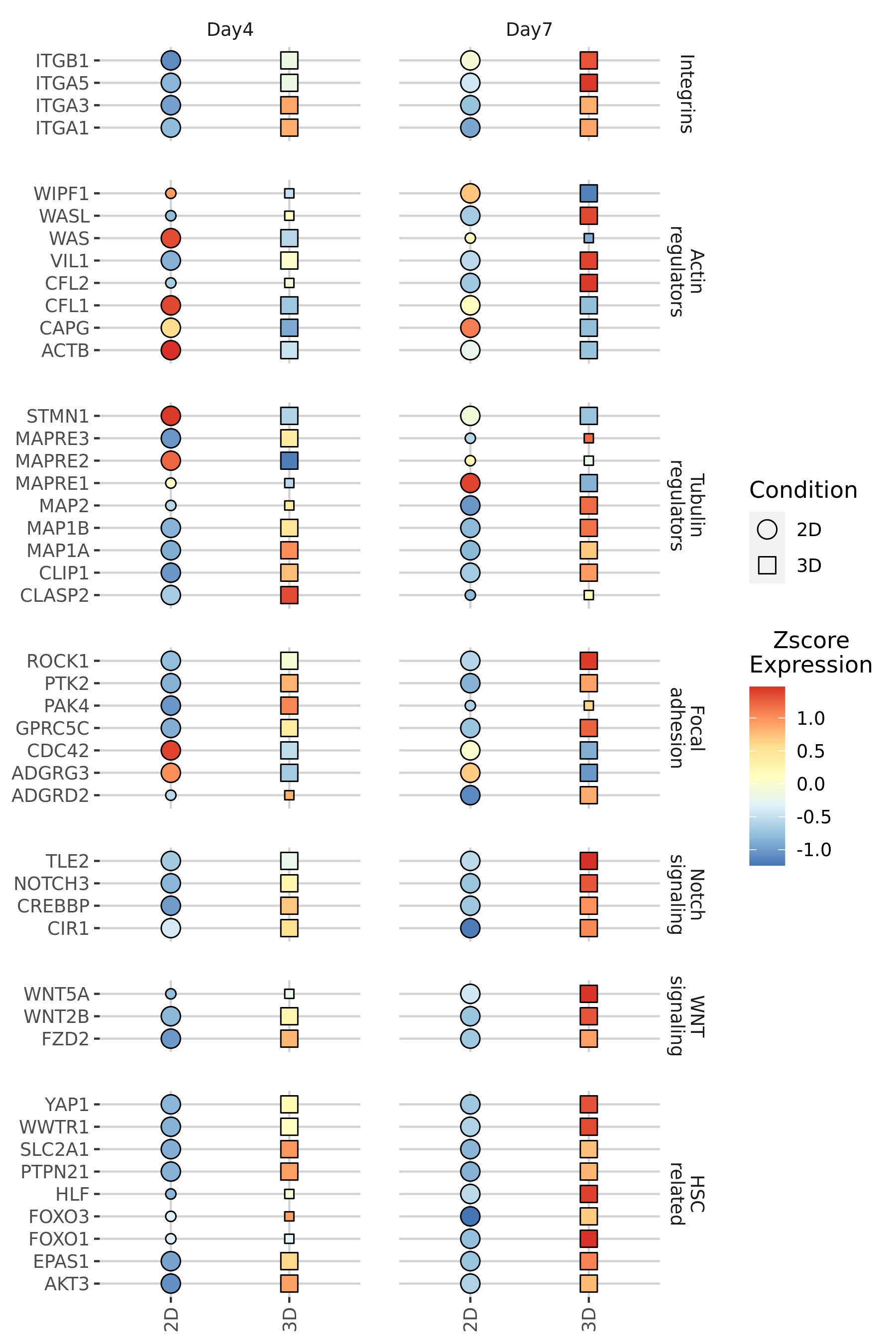
0 → 100644
350 KB
GeneList_Fig2A.xlsx
0 → 100644
File added